log rank test r package|log rank test online : makers Determine whether there are significant differences in the fitted survival distributions using a log-rank test (and/or Breslow and Tarone-Ware test). If there are differences, run a pairwise comparison post-hoc test to determine which .
webLooking for Las Vegas Hotel? 2-star hotels from $62, 3 stars from $25 and 4 stars+ from $90. Stay at The Carriage House from $132/night, The Schooner Inn from $62/night, Excalibur Hotel & Casino from $62/night .
{plog:ftitle_list}
Resultado da At SlotsUp, we provide instant access to all high-quality free slot games that can be played anytime, anywhere, as long as you’re connected to the internet. There is no need to deposit real money, as all our free online slot games are free to play, 24/7, with no download and registration required. We take .
OneSampleLogRankTest: One-Sample Log-Rank Test. The log-rank test is performed to assess the survival outcomes between two group. When there is no proper .
Description. Calculates a weighted log-rank test for the comparison of two groups. Usage. logrank.test( time, event, group, alternative = c("two.sided", "less", "greater"), rho = 0, gamma . The Log Rank test, also known as the Mantel-Haenszel test, is used to compare the survival distributions of two or more groups. This test focuses on determining if there’s a . In R, we can use the survdiff () function from the survival package to do a log-rank test, which has the following syntax: survdiff (Surv (time, status) ~ predictors, data) The Chi-Squared test statistic and related p-value are . To perform a log-rank test in R, we will use the survival package, which provides functions to perform survival analysis and the survminer package for visualization purposes. The packages can be installed using the command .
Determine whether there are significant differences in the fitted survival distributions using a log-rank test (and/or Breslow and Tarone-Ware test). If there are differences, run a pairwise comparison post-hoc test to determine which .
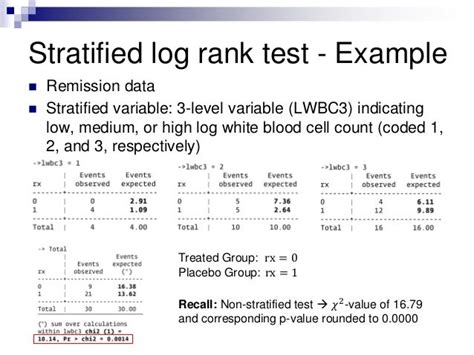
stratified log rank test
r log rank test survival
How to Perform a Log Rank Test in R, The most frequent technique to compare survival curves between two groups is to use a log-rank test. The following hypotheses are .If you actually want to measure the effects of both sex and age together on survival, you need to be doing a stratified log rank test. I've used the function SurvTest(in documentation)/surv_test . To perform a log rank test in R, we can use the survdiff() function from the survival package, which uses the following syntax: survdiff(Surv(time, status) ~ predictors, data) This function returns a Chi-Squared test statistic . rdrr.io Find an R package R language docs Run R in your browser. bgreenwell/bmisc Miscellaneous R Functions. Package index. . Sample size calculations for the log-rank test In bgreenwell/bmisc: Miscellaneous R Functions. Description Usage Arguments Details Value Note Examples. View source: R/pwr.R.
The first parameter of the Fleming-Harrington family of weighted log-rank test. Defaults to 0 for conventional log-rank test. rho2: The second parameter of the Fleming-Harrington family of weighted log-rank test. Defaults to 0 for conventional log-rank test. numSubintervals: Number of sub-intervals to approximate the mean and variance of the . Step 1: Load Necessary Packages. To perform a log-rank test in R, we will use the survival package, which provides functions to perform survival analysis and the survminer package for visualization purposes. The . As a last note, you can use the log-rank test to compare survival curves of two groups. The log-rank test is a statistical hypothesis test that tests the null hypothesis that survival curves of two populations do not differ. A certain probability distribution, namely a chi-squared distribution, can be used to derive a p-value.
In this comprehensive guide, we’ll delve deep into the application of the Log Rank test in R. 1. Introduction to the Log Rank Test. The Log Rank test, also known as the Mantel-Haenszel test, is used to compare the survival distributions of two or more groups. This test focuses on determining if there’s a statistically significant difference .The log-rank test is the most widely used method of comparing two or more survival curves. The null hypothesis is that there is no difference in survival between the two groups. . The function survdiff() [in survival package] can be used to compute log-rank test comparing two or more survival curves. survdiff() can be used as follow: surv .
r km estimator
The weight function includes Stabilized Fleming-Harrington class (rho, gamma, tau, s.tau), and any user-defined weight function. For the stabilized Fleming-Harrington class, to produce stabilized weighting function from pooled survival curve, specify either tau or s.tau, which are thresholds in survival time and survival rate, respectively. The weight function is based on . Multistate Survival Analysis using R package "Survival" 0. R: Using Log Rank Test (survdiff) 1. Logrank test for specific groups via Cox regression. 0. extract log rank (score) test result wiht p-value for Coxph Model. 4. Parameter estimates and variance for stratified variables in Cox regression (strata / survival package) 0.
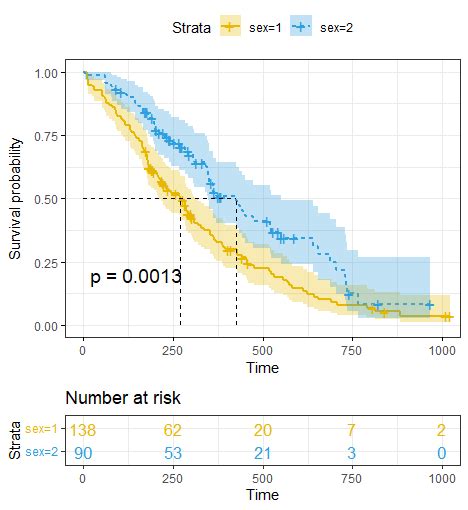
图释. 上图表示:时间对应的生存率的生存曲线图,曲线中的短竖线表示在时间点有缺失值病人。不同颜色表示不同分组,阴影部分表示95%置信区间;左下角p值表示基于log-rank test计算得到的P值,对于本图的结果p=0.0013,即不同性别的生存率有显著差异。Reference manual: lrstat.pdf : Vignettes: Comparing Direct Approximation and Schoenfeld Methods (source, R code) Sample Size Calculation With Fixed Follow-up (source, R code) Simulation for Group Sequential Trials (source, R code) Power Calculation Using Max-Combo Tests (source, R code) Sample Size Calculation Under Non-Proportional Hazards (source, R . Power of the log-rank test is estimated using simulation datasets, with user specified total sample size (in one simulation dataset), type I error, effect size, the total number of simulation datasets, sample size ratio between two comparison groups, the death rate in the reference group, and the distribution of follow-up time (simulated from a negative binomial .We would like to show you a description here but the site won’t allow us.
влагомер для дерева md918
In any case the z test statistic of each included weighted log-rank test is based on the (weighted) sum of expected minus observed events in the group corresponding to the first factor level of group. Hence a small value of the test statistic corresponds to a lower (weighted average) hazard rate in the first group.We would like to show you a description here but the site won’t allow us. This function returns a Chi-Squared test statistic and a corresponding p-value. The following example shows how to use this function to perform a log rank test in R. Example: Log Rank Test in R. For this example, . The type of weighted log-rank test. Either the default "lr" for a standard log-rank test, "mw" for a modestly-weighted log-rank test, or "fh" for the Fleming-Harrington rho-gamma family. s_star: This is a parameter for the "mw"-test. Either s_star or t_star must be specified.
The post How to Perform a Log Rank Test in R appeared first on Data Science Tutorials How to Perform a Log Rank Test in R, The most frequent technique to compare survival curves between two groups is to use a log-rank test. The following hypotheses are used in this test H0: There is no difference in survival between the two groups. HA: There is a difference in . This function is used to perform power calculation of the Log-rank test based on simulation datasets, with user specified total sample size (in one simulation), type I error, effect size, total number of simulation datasets, sample size ratio between comparison groups, the death rate in the reference group, and the distribution of follow-up time (negative binomial).The usual log-rank test is adapted to the corresponding adjusted survival curves. Rdocumentation. powered by. Learn R Programming. RISCA (version 1.0.5) Description Usage Value. Arguments. Author. Details. References. Examples Run this code .
We would like to show you a description here but the site won’t allow us. In this package we present existing methods to conduct such test named one sample log rank test. One sample log rank test for large sample sizes Approximate one sample log rank test based on Chi-squared distribution can be .
where 1 – α specifies the level of significance of statistical test and 1 – θ specifies the power of statistical test; Z α/2 and Z θ are the upper α/2 and θ percentiles of the standard normal distribution, respectively; p is the proportion of patients being assigned to the treatment arm; β 0 is the log-hazard ratio between treatment and control arms; δ is the censoring indicator (1 . We have just adopted weighted Log-rank tests to the survminer package, thanks to survMisc::comp. What are they and why they are useful? Read this blog post to find out. I used ggthemr to make the presentation a little bit more bizarre. Log-rank statistic for 2 groups Weighted Log-rank extensions Why are they useful? Plots gghtemr Log-rank (survdiff) + sea .Power of the log-rank test is estimated using simulation datasets, with user specified total sample size (in one simulation dataset), type I error, effect size, the total number of simulation datasets, sample size ratio between two comparison groups, the death rate in the reference group, and the distribution of follow-up time (simulated from a negative binomial distribution). rdrr.io Find an R package R language docs Run R in your browser. IPWsurvival . Adjusted Kaplan-Meier estimator and log-rank test with inverse probability of treatment weighting for survival data. Statistics in medicine, 24(20):3089-3110, .
Log-Rank Test for Adjusted Survival Curves. Description. The user enters individual survival data and the weights previously calculated (by using logistic regression for instance). The usual log-rank test is adapted to the corresponding adjusted survival curves. Usage ipw.log.rank(times, failures, variable, weights) Arguments The first parameter of the Fleming-Harrington family of weighted log-rank test. Defaults to 0 for conventional log-rank test. rho2: The second parameter of the Fleming-Harrington family of weighted log-rank test. Defaults to 0 for conventional log-rank test. numSubintervals: Number of sub-intervals to approximate the mean and variance of the .
predict survival time in r
30K Followers, 482 Following, 47 Posts - See Instagram photos and videos from Tiet (@therealtiiet)
log rank test r package|log rank test online